An R package for comprehensive data analysis of peptide- and protein-centric bottom-up proteomics data
A recent Bioinformatics Advances paper by the Picotti lab (Institute of Molecular Systems Biology) introduces their R package for data analysis of limited proteolysis coupled to mass spectrometry (LiP-MS) and bottom-up proteomics data.
Novel bottom-up proteomics approaches such as LiP-MS, phosphoproteomics and other PTM-centric experiments require flexible software tools to facilitate analysis and interpretation of their large and often diverse data structures. Existing R packages either offer fixed analysis pipelines or are not suited for specific user needs.
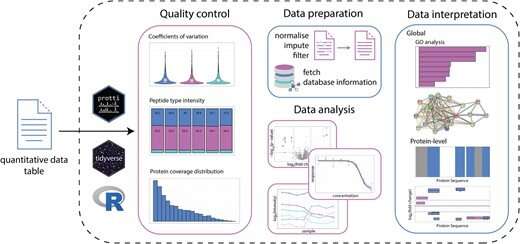
The Picotti lab introduces an R package "protti" that is user-friendly and easy to implement even for inexperienced R users. The R package is suitable for the analysis of peptide- and protein-centric data and can use various quantitative data matrices as its input. protti can be used for quality control, statistical analyses and data interpretation. While protti does not provide a graphical user interface, its design principles—based on tidyverse packages—and its documentation make it easy to understand and accessible to novice users.
Due to its flexible design, it supports analysis of label-free, data-dependent, data-independent and targeted proteomics datasets. protti can be run on the output of any search engine and software package commonly used for bottom-up proteomics experiments, such as Spectronaut, Skyline, MaxQuant or Proteome Discoverer, adequately exported to table format. Release versions are available via CRAN (https://CRAN.R-project.org/package=protti) and work on all major operating systems. The development version is maintained on GitHub (https://github.com/jpquast/protti).
More information: Jan-Philipp Quast et al, protti: an R package for comprehensive data analysis of peptide- and protein-centric bottom-up proteomics data, Bioinformatics Advances (2021). DOI: 10.1093/bioadv/vbab041
Provided by ETH Zurich